It is hard to imagine that you can estimate how
much time a cell spends in each phase of cell replication from a slide
of dead cells. Yet this is precisely what you will do in this part of the
lab. Since you are working with a prepared slide, you cannot get any information
about how long it takes a cell to divide. What you can determine is how
many cells are in each phase. From this, you can infer the percent of time
each cell spends in each phase.
Procedure
1. Observe every cell in one high power field of view and determine
which phase of the cell cycle it is in. This is best done in pairs. The
partner observing the slide calls out the phase of each cell while the
other partner records. Then switch so the recorder becomes the observer
and vice versa. Count at least two full fields of view. If you have not
counted at least 200 cells, then count a third field of view.
2. Record your data in Table 1
Table 1
|
Field 1 |
Field 2 |
Field 3 |
Total |
Percent of total cells |
Time in each stage |
| Interphase |
|
|
|
|
|
|
| Prophase |
|
|
|
|
|
|
| Metaphase |
|
|
|
|
|
|
| Anaphase |
|
|
|
|
|
|
| Telophase |
|
|
|
|
|
|
| Total cells: |
3. Calculate the percentage of cells in each phase.
Consider that it takes, on average, 24 hours (or 1,440 minutes) for
onion root-tip cells to complete the cell cycle. You can calculate the
amount of time spent in each phase of the cell cycle from the percent of
cells in that stage.
Percent of cells in stage X 1,440 minutes = minutes of cell cycle spent
in stage
Questions
1. If your observations had not been restricted to the area of the
root tip that is actively dividing, h6w would your results have been different?
2. Based on the data in Table 3.1, what can you infer about the relative
length of time an onion root-tip cell spends in each stage of cell division?
II. Crossing Over during Melosis in Sordaria
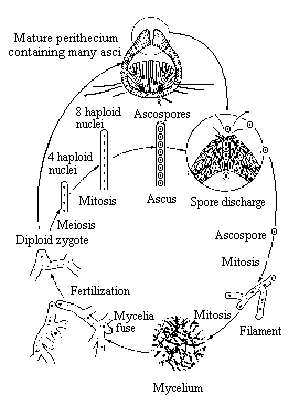
Sordaria fimicola is an ascomycete fungus
that can be used to demonstrate the results of crossing over during meiosis.
Sordaria is a haploid organism for most of its life cycle. It becomes
diploid only when the fusion of the mycelia of two different strains results
in the fusion of the two different types of haploid nuclei to form a diploid
nucleus. The diploid nucleus must then undergo meiosis to resume its haploid
state.
Meiosis, followed by mitosis, in Sordaria results
in the formation of eight haploid ascospores contained within a sac called
an ascus (plural, asci). Many asci are contained within a fruiting body
called a perithecium. When ascospores are mature the ascus ruptures, releasing
the ascospores. Each ascospore can develop into a new haploid fungus. The
life cycle of Sordaria fimicola is shown in Figure 1.
To observe crossing over in Sordaria, one
must make hybrids between wild-type and mutant strains of Sordaria.
Wild-type (+) Sordaria have black ascospores. One mutant strain
has tan spores (tn). When mycelia of these two different strains come together
and undergo meiosis, the asci that develop will contain four black ascospores
and four tan ascospores. The arrangement of the spores directly reflects
whether or not crossing over has occurred. In Figure 2, no crossing over
has occurred. Figure 3 shows the results of crossing over between the centromere
of the chromosome and the gene for ascospore color.
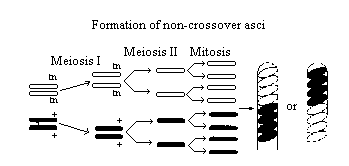
Figure 2
Two homologous chromosomes line up at metaphase I
of meiosis. The two chromatids of one chromosome each carry the gene for
tan spore color (tn) and the two chromatids of the other chromosome carry
the gene for wild-type spore color (+).
The first meiotic division (MI) results in two cells
each containing just one type of spore color gene (either tan or wild-type).
Therefore, segregation of these genes has occurred at the first meiotic
division (MI).
The second meiotic division (MII) results in four
cells, each with the haploid number of chromosomes (lN).
A mitotic division simply duplicates these cells,
resulting in 8 spores. They are arranged in the 4:4 pattern.

Figure 3
In this example, crossing over has occurred in the
region between the gene for spore color and the centromere. The homologous
chromosomes separate during meiosis I. This time, the MI results in two
cells, each containing both genes (1 tan, 1 wild-type); therefore, the
genes for spore color have not yet segregated.
Meiosis II (MII) results in segregation of the two
types of genes for spore color.
A mitotic division results in 8 spores arranged
in the 2:2:2:2 or 2:4:2 pattern. Any one of these spore arrangements would
indicate that crossing over has occurred between the gene for spore coat
color and the centromere.
Two strains of Sordaria (wild-type and tan
mutant) have been inoculated on a plate of agar. Where the mycelia of the
two strains meet (Figure 4), fruiting bodies called perithecia develop.
Meiosis occurs within the perithecia during the forrnation of asci. Prepare
a wet mount of some perithecia (the black dots in figure 4).
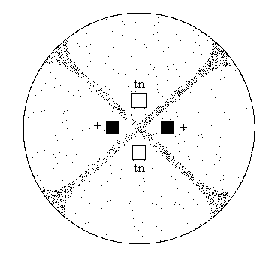
Figure 4
Gently press down on the coverslip so that the perithecia
rupture but the ascospores remain in the asci. Using the 10x objective,
view the slide and locate a group of hybrid asci (those containing both
tan and black ascospores). Count at least 50 hybrid asci and enter
your data in Table 2.
Table 2
| Number of 4:4 asci |
Number of crossover asci |
Total asci |
% showing crossover / 2 |
Gene to centromere distance |
| |
|
|
|
|
The frequency of crossing over appears to be governed
largely by the distance between genes, or in this case, between the gene
for spore coat color and the centromere. The probability of a crossover
occurring between two particular genes on the same chromosome (linked genes)
increases as the distance between those genes becomes larger. The frequency
of crossover, therefore, appears to be directly proportional to the distance
between genes.
A map unit is an arbitrary unit of measure
used to describe relative distances between linked genes. The number of
map units between two genes or between a gene and the centromere is equal
to the percentage of recombinants. Customary units cannot be used because
we cannot directly visualize genes with the light microscope. However,
due to the relationship between distance and crossover frequency, we may
use the map unit.
Analysis of Results
1. Using the data in Table 2, determine the distance between the gene
for spore color and the centromere. Calculate the percent of crossovers
by dividing the number of crossover asci (2:2:2:2 or 2:4:2) by the total
number of asci x 100. To calculate the map distance, divide the percentage
of crossover asci by 2. The percentage of crossover asci is divided by
2 because only half of the spores in each ascus are the result of a crossover
event (Figure 3). Record your results in Table 2.
2. Draw a pair of chromosomes in MI and MII, and show how you would
get a 2:4:2 arrangement of ascospores by crossing over. (Hint: refer to
Figure 3).



